Development of TRPML1-4A assays across manual, automated patch-clamp, and fluorescence-based platforms
Case Study
Case Study title
Development of TRPML1-4A assays across manual, automated patch-clamp, and fluorescence-based platforms
Authors
Molly Rowlett, Thomas Hill, Scott Maidment, Luke Marrington, Ayesha Jinat, Alexandra Pinggera, Katie Puddefoot, Gary Clark, Zeki Ilkan, Robert Kirby, Graham Smith
Metrion Biosciences Ltd, Granta Centre, Granta Park, Cambridge, CB21 6AL, United Kingdom
Introduction
TRPML1 (transient receptor potential mucolipin subfamily 1) is a non-selective cation channel primarily localised in late endosomes and lysosomes. Channel activity is regulated by both calcium (Ca2+) and pH. TRPML1 facilitates Ca2+ efflux from the lysosomal lumen and is, therefore, vital for vesicular trafficking processes such as exocytosis and autophagy1.
Mutations in the gene encoding the channel protein (MCOLN1) results in lysosomal storage diseases, such as Mucolipidosis type IV (ML4) which presents as motor deficits, neurodegeneration and visual impairments2.
The development and validation of electrophysiological assays to study TRPML1 is important to understand the function and pharmacology of the channel. Here, we used a TRPML1 variant that lacks the endo-lysosomal retention sequences (TRPML1-4A), enabling the channel to express at the plasma membrane3. As such channel behaviour can be characterised by means of whole-cell patch-clamp and fluorescence-based techniques.
Aim
To develop a suite of screening assays using manual patch-clamp, automated patch-clamp and fluorescence-based platforms capable of identifying modulators of the TRPML1-4A channel.
Materials and methods
Cell culture
- TRPML1 channel sequence contains two di-leucine sorting motifs that are responsible for endo-lysosomal trafficking. Mutations at these sites gives rise to the -4A mutant, which allows accumulation of TRPML1 at the plasma membrane3.
- An inducible monoclonal human embryonic kidney 293 (HEK293) cell line expressing the TRPML1-4A variant (HEK-TRPML1-4A) was generated by Metrion’s cell biology team.
- TRPML1 channel expression was induced by the addition of doxycycline into culture media (0.25 µg/ml to 1 µg/ml), 24 to 48 hours prior to experiments.
Compounds
- TRPML1-4A currents were elicited using the agonist, ML-SA5 (Alomone Labs), stocks of which were prepared in 100% DMSO to 30 mM. Test concentrations were prepared in the appropriate solution each experimental day to a final DMSO concentration of either 0.1% or 0.3% v/v.
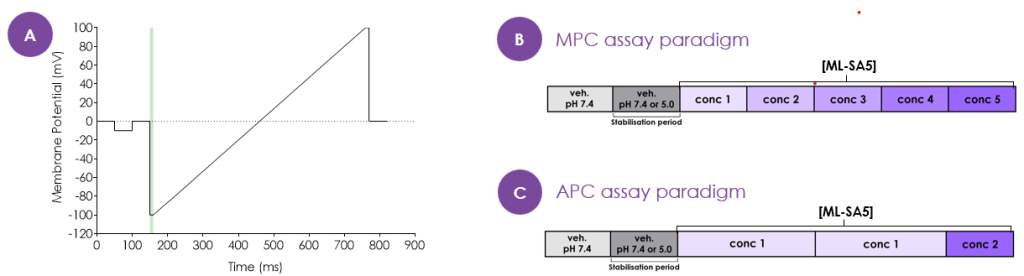
Patch-clamp protocol and paradigms
- Manual patch-clamp recordings were performed on a Multiclamp 700B and 1400A digitiser device (Molecular Devices) and automated patch-clamp recordings were performed on Qube 384 (Sophion).
- The following voltage protocol and assay paradigms were utilised in patch-clamp recordings:
Patch-clamp solutions
The intracellular (ICS) and extracellular solutions (ECS) used in patch-clamp assays are summarised below:
ICS (in mM); CsF 120, NaCl 15, CaCl2 1, MgCl2 1, HEPES/Citric acid 10, EGTA 10, pH 7.2.
ECS for MPC (in mM); NMDG 100, NaCl 35, KCl 5, CaCl2 0.5, MgCl2 1, HEPES/Citric acid 10, Glucose 11.1, pH 7.4/5.
ECS for APC (in mM); NaCl 140, KCl 13, CaCl2 1, MgCl2 1, BaCl2 4, HEPES/MES 10, pH 7.4/5.
Fluorescence-based assay
- Fluorescence-based measurements were performed on a FLIPR Penta platform (Molecular Devices).
- Hanks buffered salt solution (HBSS + Ca2+/Mg2+) was supplemented with HEPES (20 mM) to pH 7.4.
- HEK-TRPML1-4A cells were incubated with FLIPR calcium 6 assay dye (Molecular Devices) for 1 hour prior to the experiment.
- Fluorescent response was measured for 3 minutes after compound addition following a basal read of 20 seconds.
Results
1. Validation of the TPRML1-4A stable cell line using manual patch-clamp technique

2. Development of an assay for use on the automated patch-clamp platform, Qube 384

3. Development of a fluorescence-based assay using the FLIPR platform
The reference agonist, ML-SA5 was tested at 10 concentrations (0.0003 to 10 µM) at pH 7.4.
Two assay runs were performed to ensure reproducibility between experiments.
Area under curve was calculated for each well and concentration-response curves were constructed.

Conclusions
- We successfully generated a HEK-TRPML1-4A stable cell line which can be functionally characterised using electrophysiological techniques.
- Leveraging both patch-clamp electrophysiology and fluorescence-based assays, we were able to optimise screening services capable of studying TRPML1 modulators with excellent cross-platform correlation.
- The EC50 values we obtained for ML-SA5 were in line with literature for FLIPR5 and Qube6 platforms.
- The pH sensitivity of TRPML1 was confirmed in our patch clamp assays showing greater response to ML-SA5 in acidic pH conditions.
References
- 1.Li G. et al., Adv Exp Med Biol. 2021, PMID: 35138619
- 2.Wu G. et al., J Biol Chem. 2018, PMID: 29884771
- 3.Vergarajauregui S, et al., Traffic. 2006, PMID: 16497227
- 4.Baracaldo-Santamaria D. et al., Int J Mol Sci, 2023, PMID: 37240413
- 5.Peng X. et al., Bioorganic & Medicinal Chemistry Letters, 2024, PMID: 38141860
- 6.Sauter D. et al., Sophion Bioscience (presented at BPS. 2023)

Let’s work together
What are your specific ion channel and assay needs?
If you have any questions or would like to discuss your specific assay requirements, we will put you directly in touch with a member of our scientific team. Contact us today to discover more.
